 https://ibmm.umontpellier.fr/wp-content/uploads/2025/06/Morgan.jpg
226
400
Caroline Clavel
https://ibmm2.umontpellier.fr/wp-content/uploads/logo_ibmm_hd.jpg
Caroline Clavel2025-06-19 16:40:542025-06-19 16:51:18Morgan Pellerano présente son métier sur le site de l’UM
https://ibmm.umontpellier.fr/wp-content/uploads/2025/06/Morgan.jpg
226
400
Caroline Clavel
https://ibmm2.umontpellier.fr/wp-content/uploads/logo_ibmm_hd.jpg
Caroline Clavel2025-06-19 16:40:542025-06-19 16:51:18Morgan Pellerano présente son métier sur le site de l’UM- En savoir +
L’Institut des Biomolécules Max Mousseron est une Unité Mixte de Recherche (UMR 5247) UM, CNRS, ENSCM qui développe des recherches à l’interface de la chimie, de la biologie et de la santé intégrant les enjeux du développement durable : nourrir, soigner, protéger.

Actualités
 https://ibmm.umontpellier.fr/wp-content/uploads/2025/06/Morgan.jpg
226
400
Caroline Clavel
https://ibmm2.umontpellier.fr/wp-content/uploads/logo_ibmm_hd.jpg
Caroline Clavel2025-06-19 16:40:542025-06-19 16:51:18Morgan Pellerano présente son métier sur le site de l’UM
https://ibmm.umontpellier.fr/wp-content/uploads/2025/06/Morgan.jpg
226
400
Caroline Clavel
https://ibmm2.umontpellier.fr/wp-content/uploads/logo_ibmm_hd.jpg
Caroline Clavel2025-06-19 16:40:542025-06-19 16:51:18Morgan Pellerano présente son métier sur le site de l’UM
Grand succès pour le congrès Mech’cheM !
Grand succès pour le congrès Mech'cheM qui s'est déroulé…
16 équipes de recherche
Polymères pour la Santé et Biomatériaux
10 janvier 2024/par Stephanie HERNANDEZ
Chimie Verte et Technologies Innovantes
10 janvier 2024/par Stephanie HERNANDEZ
Oncopharmacochimie & Pharmacotoxicologie Cutanée
10 janvier 2024/par Stephanie HERNANDEZ
Oncothérapie et oncopharmacologie
10 janvier 2024/par Stephanie HERNANDEZ
Dynamique des Systèmes Biomoléculaires Complexes
10 janvier 2024/par Stephanie HERNANDEZ
Glyco et Nanovecteurs pour le ciblage Thérapeutique
10 janvier 2024/par Stephanie HERNANDEZ
Sciences analytiques des biomolécules
10 janvier 2024/par Stephanie HERNANDEZ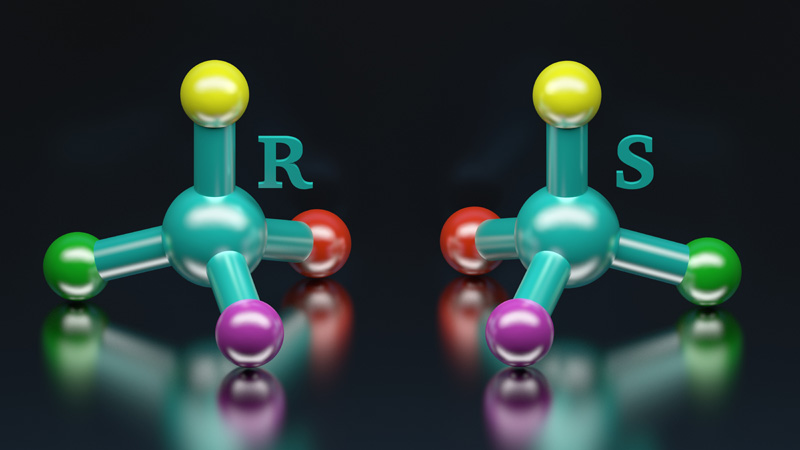
Synthèse stéréosélective & acides aminés modifiés
10 janvier 2024/par Stephanie HERNANDEZ
Acides aminés, hétérocycles, peptides & protéines
10 janvier 2024/par Stephanie HERNANDEZ
Pharmacologie Cellulaire
10 janvier 2024/par Stephanie HERNANDEZ
ChemBioNAC
10 janvier 2024/par Stephanie HERNANDEZ
Pharmacochimie, transmission synaptique & neuroprotection
10 janvier 2024/par Stephanie HERNANDEZ
Supramolecular MAchines and ARchitecture Team
10 janvier 2024/par Stephanie HERNANDEZ
Synthèse de Lipides Bioactifs
10 janvier 2024/par Stephanie HERNANDEZ
Nucléosides & Effecteurs Phosphorylés
10 janvier 2024/par Stephanie HERNANDEZ







Offre de thèse : PHOTOCLICK: Development of biorthogonal photo-crosslinking reactions for 3D printing micro-structured biomaterials
Contract: 3 years starting from October 2025 Application deadline: 04/07/2025 Location: Institute of Biomolecules Max Mousseron (IBMM), Montpellier, France and Institute for Molecular Systems Engineering and Advanced Materials (IMSEAM), Heidelberg, Germany The project This thesis project is located at the interface of click chemistry, photochemistry, materials, 3D printing and biology. The goal is to […]
Offre de thèse : Advanced 4D biomaterials for mucosa and sub-mucosa treatment in patients affected by intestinal diseases (Daedalus)
ADVANCED 4D BIOMATERIALS FOR MUCOSA AND SUB-MUCOSA TREATMENT IN PATIENTS AFFECTED BY INTESTINAL DISEASES (Daedalus) financed by the EU program HORIZON EUROPE 4D biomaterials enable new surgical treatments by autonomously acting in response to environmental stimuli, thus overcoming the limitations of standard medical strategies. DAEDALUS is a European project that gathers 13 partners and […]